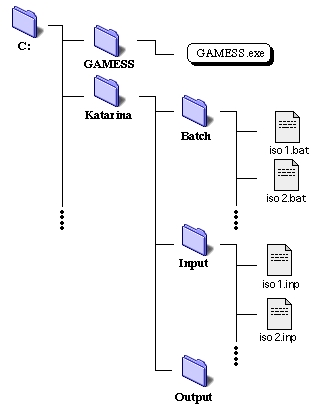
 | Figure 1. File tree of necessary files for GAMESS run |
|---|
The structure of file tree necessary for a GAMESS run on a compute node for our Condor-NT system at this site is shown in Figure 1. The only requirement for a compute node is to have a copy of GAMESS executable and a user folder (directory), here we call it, "Katarina", containing folders, Batch, Input, and Output. Before job submission, all of the batch files and input files created must be place in appropriate places using a batch script called copy.bat, shown in the cheap compuating page.
 | Figure 1. File tree of necessary files for GAMESS run |
|---|
Once a job is submitted to the compute node, the batch file in the Batch directory is executed. The content of the batch file is also shown in the cheap computing page. The batch file looks for a corresponding input file, and execute GAMESS program. The output files are placed into the Output directory.
We typically use Chem3D to draw 3-dimensional structures first. We further optimize the geometry of the molecule by MM2 or MOPAC within Chem3D. The optmized geoemtry is saved as Cartesian coordinate, usually with an option "missing"; without all connectivity and atom type information (Chem3D puts xxx.cc1 extension on each file). The geometries of many conformers are generated in this fashon, and saved in a directory.
The GAMESS input files are generated by extracting the geometry data saved in xxx.cc1 files. We prepare input files by using the following csh script, called mkinp, under Cygwin. We will show in the next section a modification to this script to prepare an input file with added isomer and protonation numbers in the input by using sed, and prepare corresponding batch and submission files.
#!/bin/tcsh # set ver = $1.cc1 # awk -f extrct_geom < $ver >> temp # cp template $ver.log cat temp >> $ver.log cat endfile >> $ver.log mv $ver.log $ver.inp # exit |
Using tcsh |
| First argument from command line ($1) ($ver) | |
| Awking to extract geometry info from $1.cc1 | |
| Copy a GAMESS template into $ver.log | |
| Append geometry extracted into $ver.log | |
| Append $end into $ver.log | |
| Rename $ver.log to $ver.inp | |
| Exit the program |
The awk script used in the above csh script looks like the following
NF>=4 {
if($1 == "C") { chrg = "6.0" }
else if($1 == "H") { chrg = "1.0" }
else if($1 == "O") { chrg = "8.0" }
else if($1 == "N") { chrg = "7.0" }
print $1 " " chrg " " $2 " " $3 " " $4
}
|
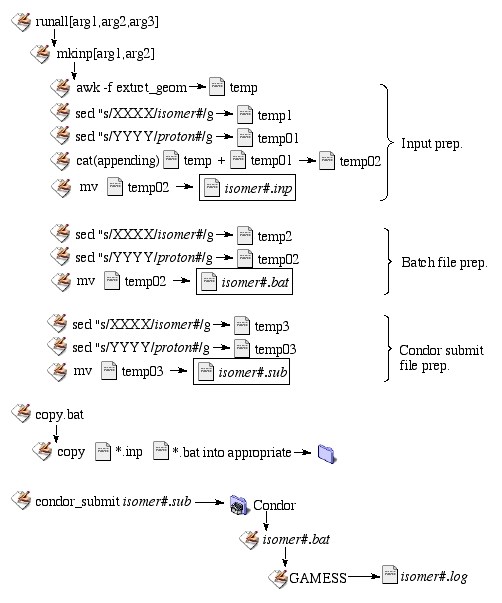
By applying the scripts above, one can make little more fancier scripts to do the input and other file preparation. The following fiigure explains the steps involved in input preparation and job submission for protonated molecules which need more than one designation for the location of the proton and isomer number. A series of scripts are executed to prepare input files, as explained above and other page. Subsequently, the files are copied and submitted to Condor queue.
 |
 |
| Figure 2. Flow chart of input prep. to job submission. |
The csh script, mkinp, takes two arguments; first being which isomer and second being protonation site (if needed). We can further automate the input preparation by using runall csh script, which asks for three arguments. First argument is which isomer to start, and the second is for which isomer to end. The third one (if needed) is a protonation site.
Subsequently, the batch script is prepared by mkinp. Only the isomer number, which is part of input file name and the protonation site designation is changed from a template. Preparing a job submission file needed by Condor is edited the same way as the isomer number by mkinp.
Once all input, batch and submission files are created, the input and batch files are copied into appropriate directories in "Katarina" directory using copy.bat on all compute nodes.
Finally, the jobs are submitted to Condor by using "condor_submit" command (see below). The queue system looks for an idle computer and submit a batch file (isomer#.bat) to the compute node. GAMESS is executed and a log file is created in an "Output" directory.
Condor commands. There is only a few commands necessary for submission and checking queue on Condor.
| Command | what it does |
|---|---|
| condor_status | checking which cocmputers are up |
| condor_q | submitted jobs from the current console |
| condor_submit isomer#.sub | submitting isomer#.sub into a queue |
| condor_rm pid | delete a job with pid |
Availabe computers. Currently, we have 9 computers in the cluster at Science Division Computer Facility (SDCF). All computers are set up for GAMESS run. We can easily adapt to any other types of jobs such as Gausssian98W. The available computers are shown below.
50. -DELL-01- /home/Administrator> condor_status
Name OpSys Arch State Activity LoadAv Mem ActvtyTime
23mya.liunet. WINNT40 INTEL Owner Idle 1.003 128 0+00:25:48
23mzu.liunet. WINNT40 INTEL Unclaimed Idle 0.033 128 0+00:05:04
23n1a.liu.edu WINNT40 INTEL Unclaimed Idle 1.520 128 0+01:01:05
23noe.liunet. WINNT40 INTEL Owner Idle 0.004 128365+00:06:23
dell-01.liune WINNT40 INTEL Unclaimed Idle 0.032 127 0+00:07:42
dell-02.liune WINNT40 INTEL Unclaimed Idle 0.003 127 0+00:08:28
dell-03.liune WINNT40 INTEL Claimed Busy 0.956 127 0+00:05:57
dell-04.liune WINNT40 INTEL Unclaimed Idle 0.001 127 0+01:10:25
dell-05.liune WINNT40 INTEL Unclaimed Idle 0.002 127 0+01:09:11
Machines Owner Claimed Unclaimed Matched Preempting
INTEL/WINNT40 9 2 1 6 0 0
Total 9 2 1 6 0 0
|
More to come!
Nikita Matsunaga 7/18/01